We are group of people dedicated to Bioinformatics research and service provision.
Within the fields of bioinformatics and systems biology, we seek to apply Big-Data technologies and methods to answer questions in plant biology, specifically in studies of crop genomics and genetics.
The main research interests include:
Artificial intelligence (AI) and Big-Data analysis,
Epitranscriptome: Bioinformatics approaches and regulation mechanisms,
Bioinformatics-based genotype-phenotype mapping,
Systems biology approaches-based plant stress research.
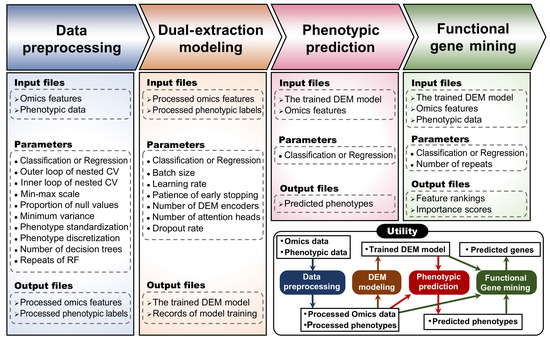
A multi-modal deep-learning architecture designed to extract representative features from heterogeneous omics datasets, enabling the …
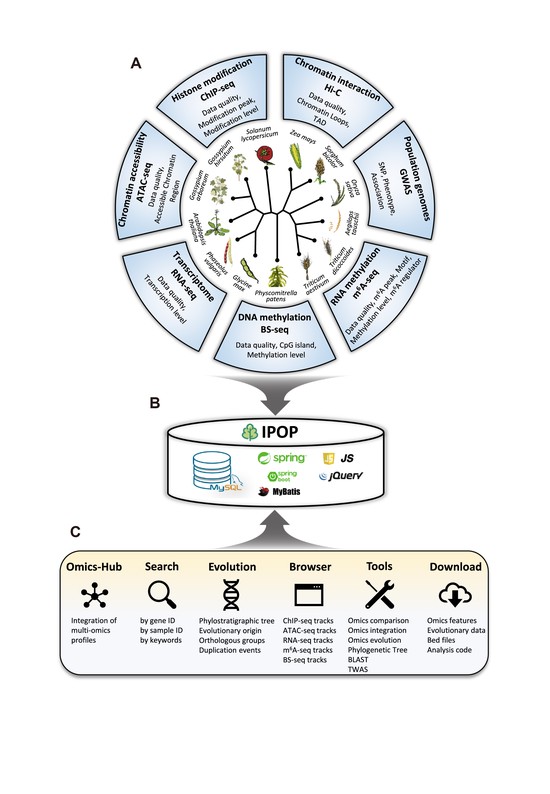
An Integrative Plant Multi-omics Platform for Cross-species Comparison and Evolutionary Study
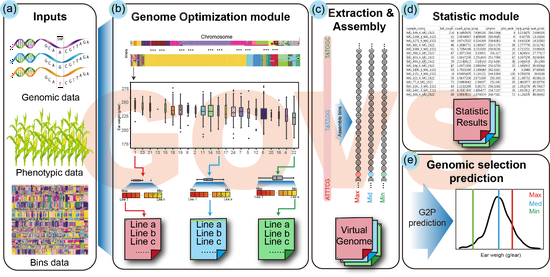
Genome optimization via virtual simulation to accelerate maize hybrid breeding

A Database for Small Molecules with Functional Implications in Plants
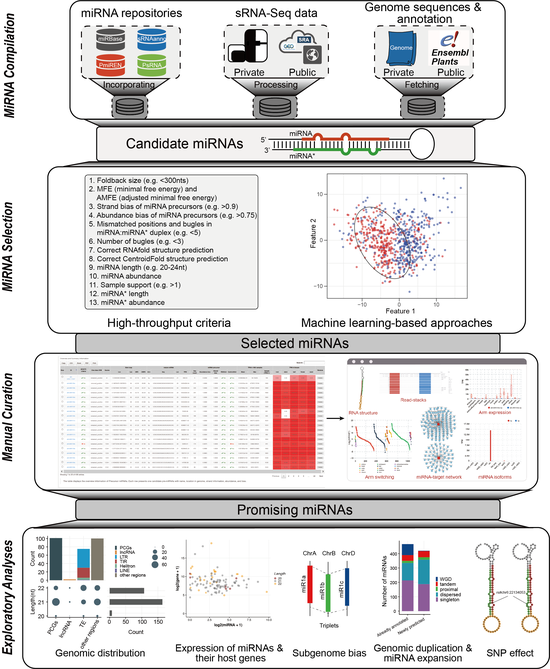
Interactive Web-based Annotation of Plant MicroRNAs with iwa-miRNA
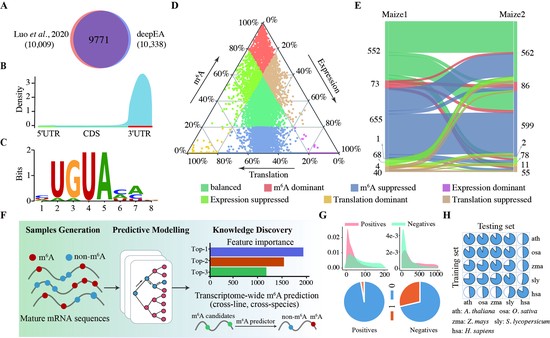
deepEA a containerized web server for interactive analysis of epitranscriptome sequencing data
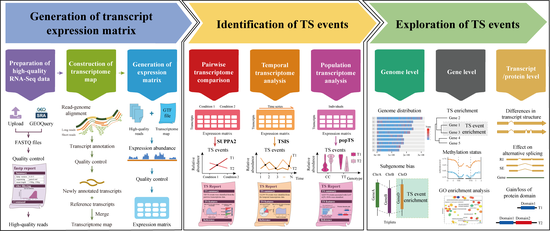
Exploring transcriptional switches from pairwise, temporal, and population RNA-Seq data using deepTS
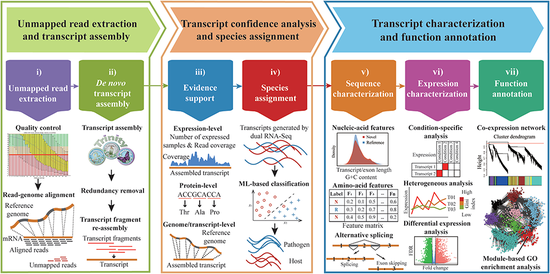
A bioinformatics framework for comprehensive assembly and functional annotation
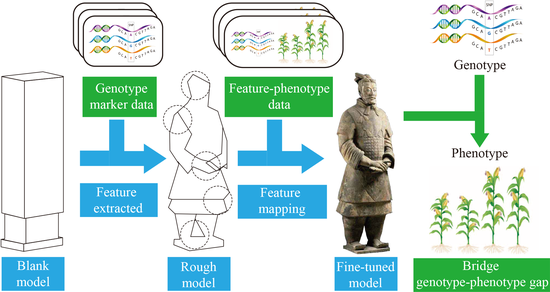
Deep learning-based genomic selection
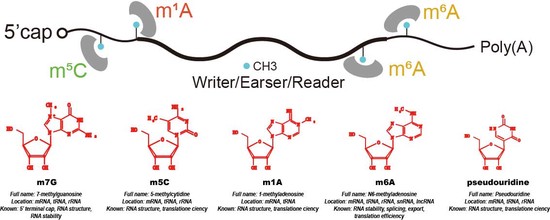
Bioinformatics approaches for epitranscriptome analysis and their applications
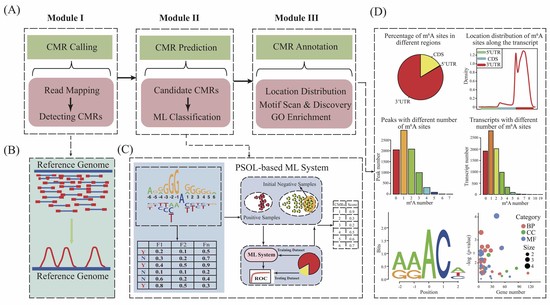
Plant epitranscriptome analysis
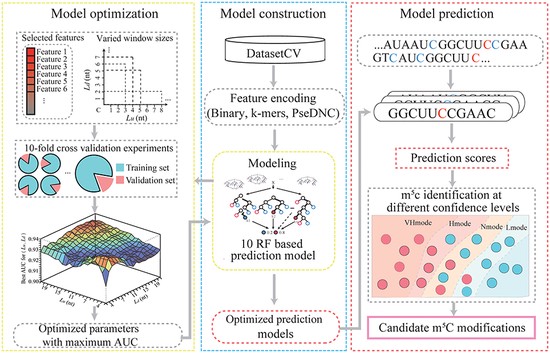
Machine learning-based m5C prediction
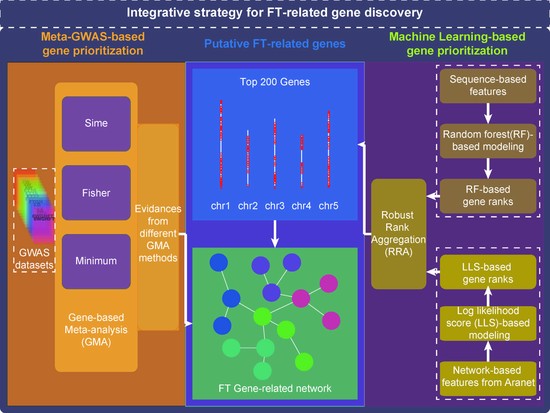
Bioinformatics approaches for genotype-to-phenotype mapping and their applications
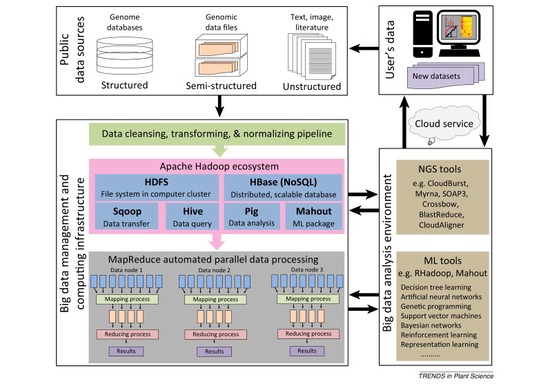
Artificial intelligence (AI)-based Big-data analysis approaches and their applications
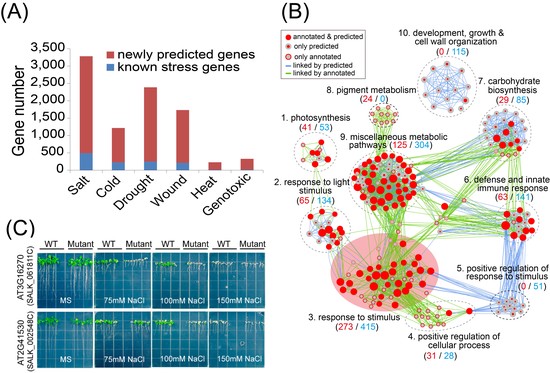
Machine learning-based differential network analysis
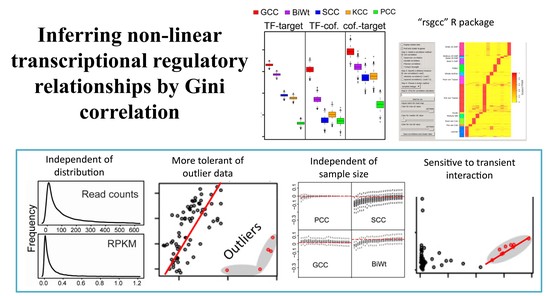
Gini-based transcriptiome analysis
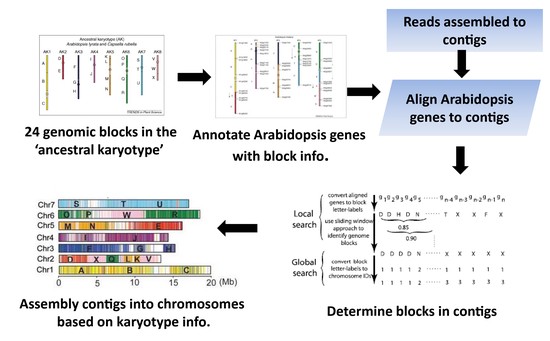
Karyotype-based genome assembly for Brassicaceae species