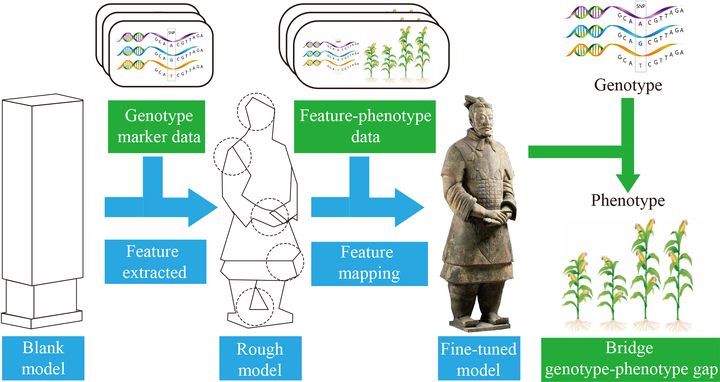
Genomic selection (GS) is a promising breeding strategy by which the phenotypes of plant individuals are usually predicted based on genome-wide markers of genotypes. Conventional GS models typically make strong assumptions, resulting in difficulty capturing complex relationships within genotypes and between genotypes and phenotypes. To address this problem, we presented a deep learning (DL)-based model, named DeepGS, to automatically learn complex relationships without pre-defined rules. The experimental results indicate that DeepGS can be used as a complement to the commonly used RR-BLUP for more accurately selecting individuals with high phenotypic values.
DeepGS software: DeepGS is an R package for predicting phenotypes from genotypes using a deep convolutional neural network approach.
Figure. Pipeline of the Predicting Phenotypes from Genotypes by DeepGS.
- View Details
- Please cite the following article:
Ma, W., Qiu, Z., Song, J., Li, J., Cheng, Q., Zhai, J., Ma, C. (2018). A deep convolutional neural network approach for predicting phenotypes from genotypes Planta 248(5), 1307-1318. https://dx.doi.org/10.1007/s00425-018-2976-9