KGBassembler
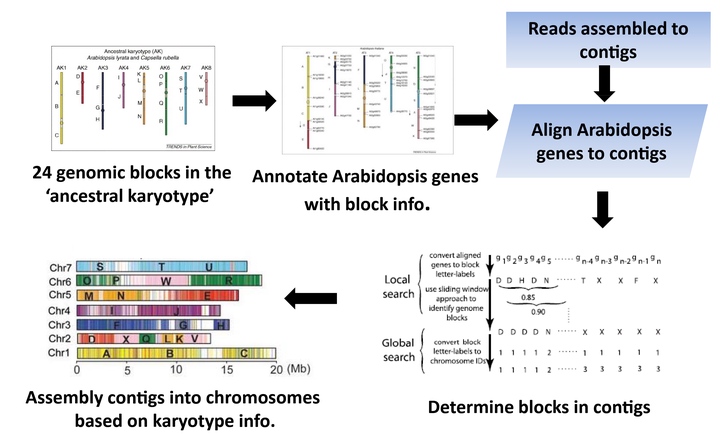
The Brassicaceae family includes the most important plant model Arabidopsis thaliana and many cruciferous vegetable crops. Due to the lack of genetic and/or physical maps for many non-model Brassicaceae species, de novo assembly of full chromosomes is difficult in many experiments designed with short sequencing data. To address this problem, we developed a bioinformatics tool named KGBassembler to automatically finalize assembly of full chromosomes from contigs and/or scaffolds based on available karyotypes of Brassicaceae species (Figure 1). KGBassembler has been successfully applied to the assembly of Eutrema salsugineum, an extremophile model for abiotic stress tolerance studies.
- View Details
- Please cite the following article:
Ma, C., Chen, H., Xin, M., Yang, R., Wang, X. (2012). KGBassembler: a karyotype-based genome assembler for Brassicaceae species Bioinformatics 28(23), 3141-3143. https://dx.doi.org/10.1093/bioinformatics/bts586