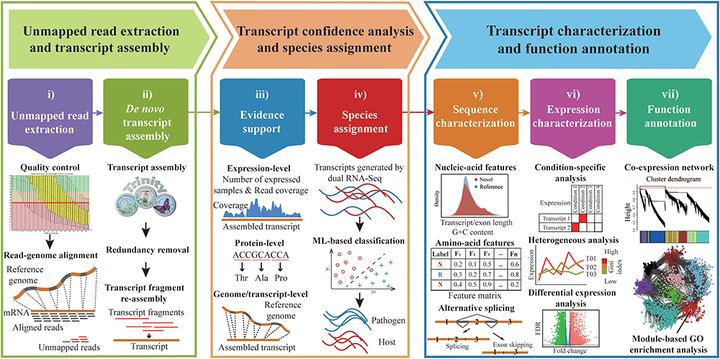
CAFU is a Galaxy-based bioinformatics framework for comprehensive assembly and functional annotation of unmapped RNA-seq data from single- and mixed-species samples which integrates plenty of existing next-generation sequencing (NGS) analytical tools and our developed programs, and features an easy-to-use interface to manage, manipulate and most importantly, explore large-scale unmapped reads. Besides the common process of reads cleansing and mapping, unmapped reads extraction and de novo transcription assembly, CAFU optionally offers multiple-level evidence evaluation, sequence and expression characterization, and transcript function annotation. Taking advantages of machine learning (ML) technologies, CAFU also effectively addresses the challenge of classifying species-specific transcripts assembled using unmapped reads from mixed-species samples.
- Web Server
- View Details
- Please cite the following article:
Chen, S., Ren, C., Zhai, J., Yu, J., Zhao, X., Li, Z., Zhang, T., Ma, W., Han, Z., Ma, C. (2019). CAFU: a Galaxy framework for exploring unmapped RNA-Seq data Briefings in Bioinformatics https://dx.doi.org/10.1093/bib/bbz018