miRLocator: Machine Learning-Based Prediction of Mature MicroRNAs within Plant Pre-miRNA Sequences
Abstract
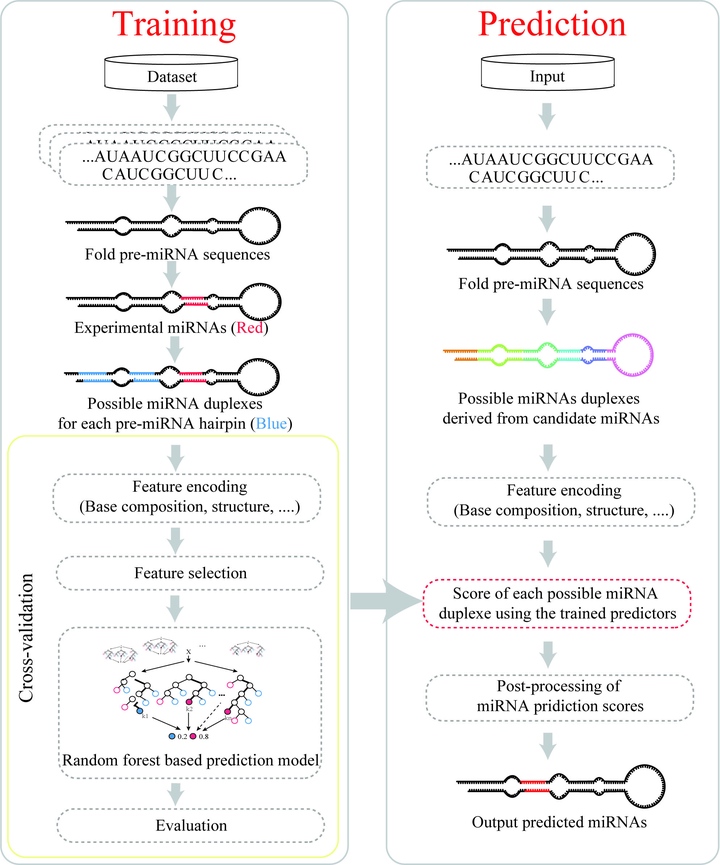
MicroRNAs (miRNAs) are a class of short, non-coding RNA that play regulatory roles in a wide variety of biological processes, such as plant growth and abiotic stress responses. Although several computational tools have been developed to identify primary miRNAs and precursor miRNAs (pre-miRNAs), very few provide the functionality of locating mature miRNAs within plant pre-miRNAs. This manuscript introduces a novel algorithm for predicting miRNAs named miRLocator, which is based on machine learning techniques and sequence and structural features extracted from miRNA:miRNA duplexes. To address the class imbalance problem (few real miRNAs and a large number of pseudo miRNAs), the prediction models in miRLocator were optimized by considering critical (and often ignored) factors that can markedly affect the prediction accuracy of mature miRNAs, including the machine learning algorithm and the ratio between training positive and negative samples. Ten-fold cross-validation on 5854 experimentally validated miRNAs from 19 plant species showed that miRLocator performed better than the state-of-art miRNA predictor miRdup in locating mature miRNAs within plant pre-miRNAs. miRLocator will aid researchers interested in discovering miRNAs from model and non-model plant species.