Exploring transcriptional switches from pairwise, temporal, and population RNA-Seq data using deepTS
Abstract
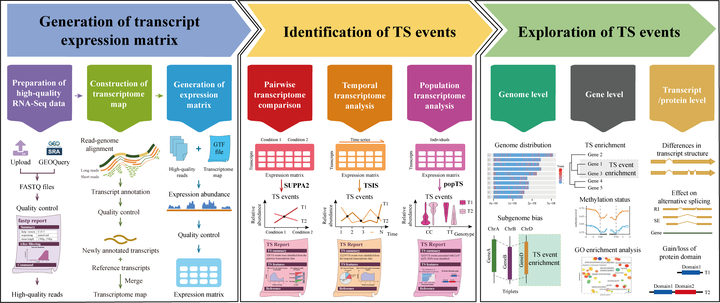
Transcriptional switch (TS) is a widely observed phenomenon caused by changes in the relative expression of transcripts from the same gene, in spatial, temporal, or other dimensions. TS has been associated with human diseases, plant development, and stress responses. Its investigation is often hampered by a lack of suitable tools allowing comprehensive and flexible TS analysis for high-throughput RNA sequencing (RNA-Seq) data. Here, we present deepTS, a user-friendly web-based implementation that enables a fullyinteractive, multifunctional identification, visualization and analysis of TS events for large-scale RNA-Seq data sets from pairwise, temporal, and population experiments. deepTS offers rich functionality to streamline RNA-Seq-based TS analysis for both model and non-model organisms and for those with or without reference transcriptome. The presented case studies highlight the capabilities of deepTS and demonstrate its potential for the transcriptome-wide TS analysis of pairwise, temporal, and population RNA-Seq data. We believe deepTS will help research groups, regardless of their informatics expertise, perform accessible, reproducible, and collaborative TS analyses of large-scale RNA-Seq data.