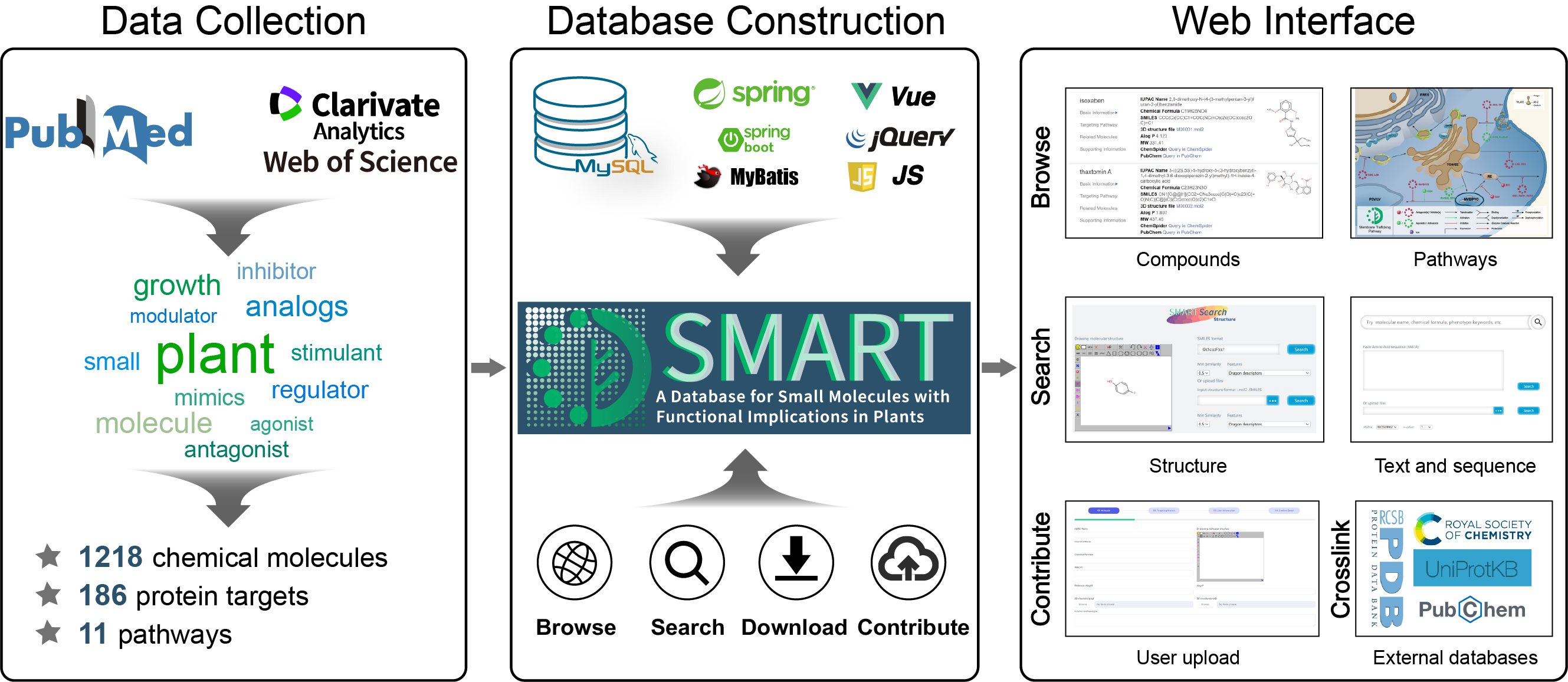
Introduction
SMART is a database for small molecules with functional implications in plants. In the current version, the SMART database provides 1218 small molecules with experimentally realized function information in plants. These 1218 small molecules are involved in 10 biological pathways and associated with 186 proteins, 108 of these 1218 small molecules are experimentally confirmed to target 24 proteins, exhibiting agonizing, antagonizing or inhibiting activity.
Browse
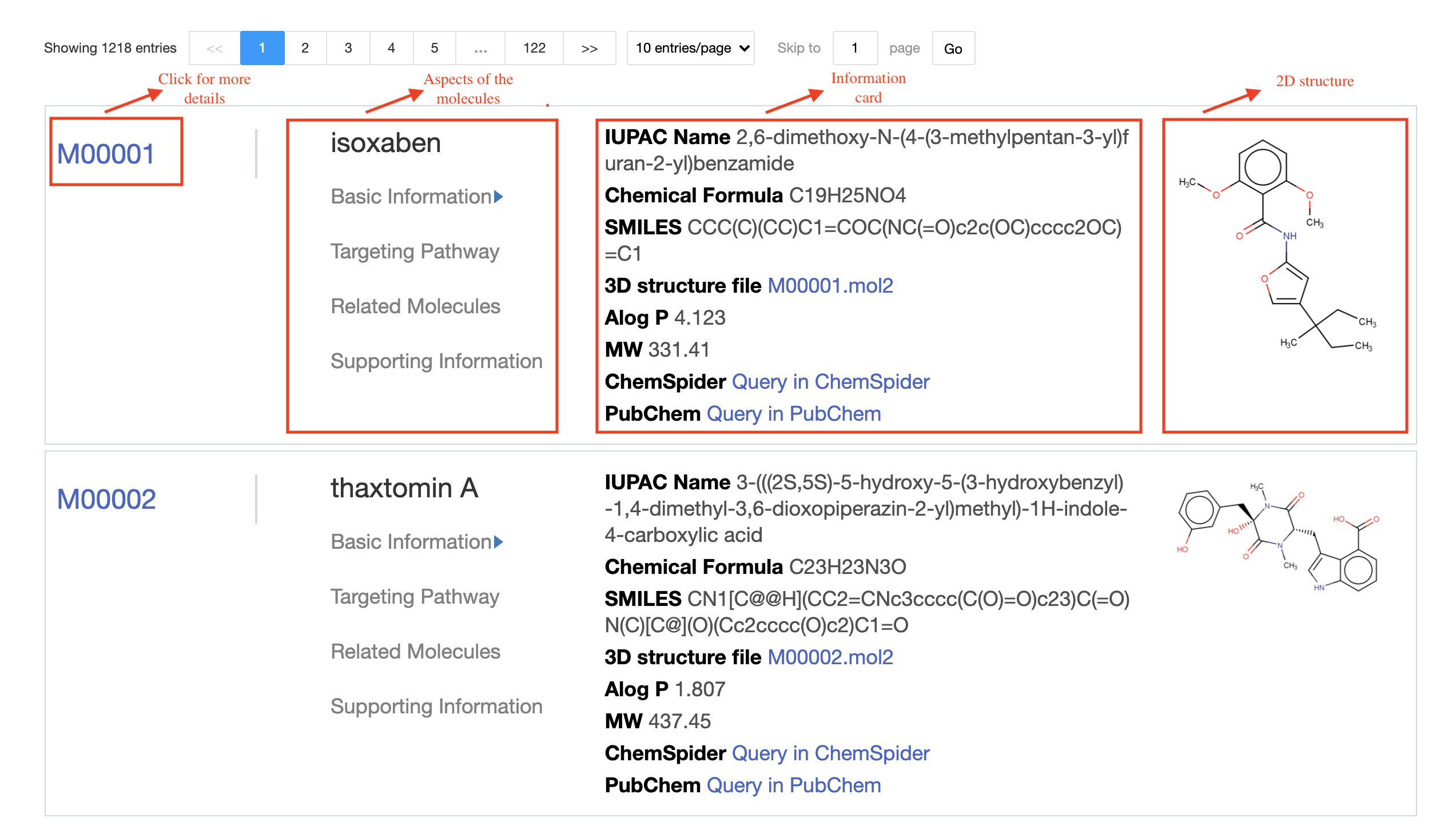
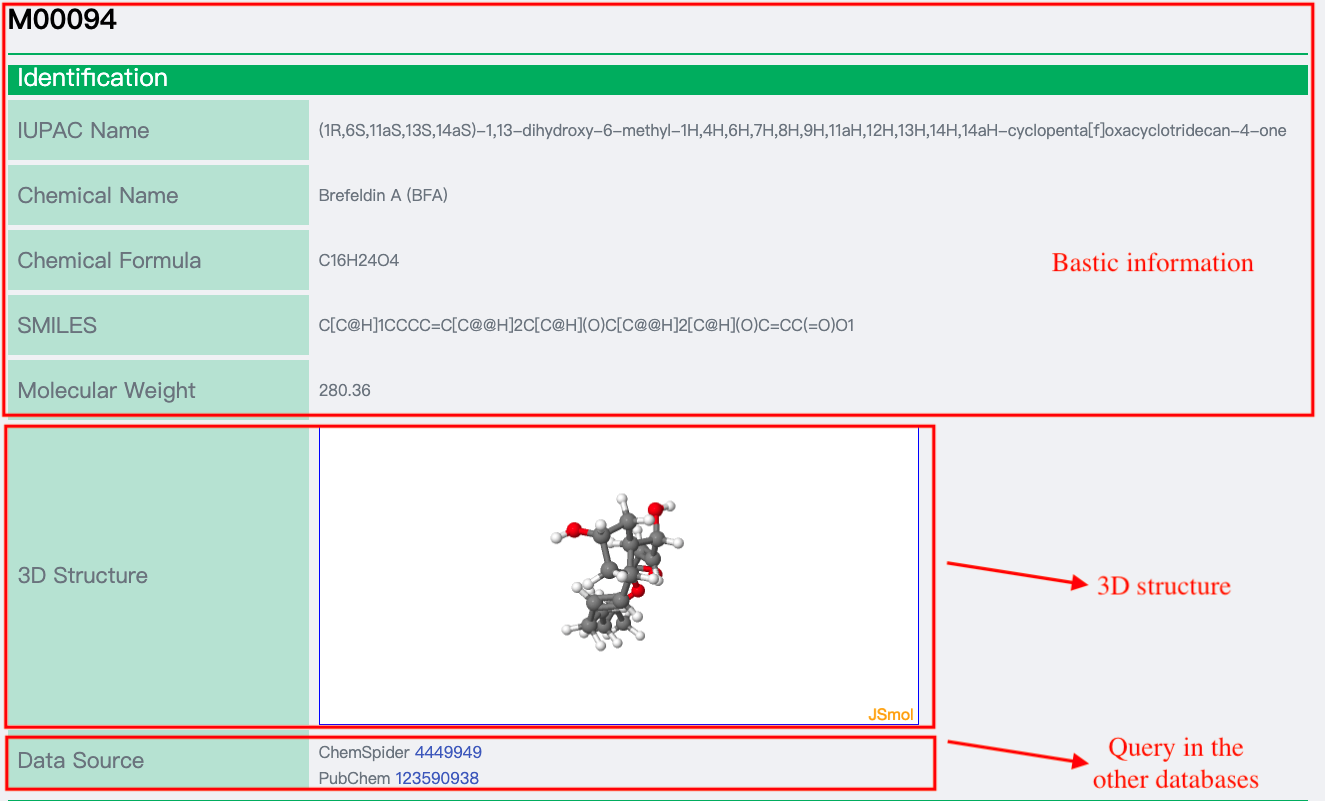
SMART provides three ways(Molecule, Pathway and Target) to browse the SMART database. On the Browse Molecule page, the information of the four aspects of the molecule is displayed, including Basic Information, Pathway, Related Compounds and Supporting information. In the current version of SMART, related molecules were defined as active molecules tested in a single literature or molecules targeting on the same protein targets.You can switch between each other by clicking the title on the information card, and click elsewhere on the card can browse the molecular detailed information page. The Browse Pathway page shows different plant signal pathways, click on the molecular icons can enter the corresponding page. The Browse Target page is presented as a table of protein information.
Browse-Molecule

Browse-Pathway
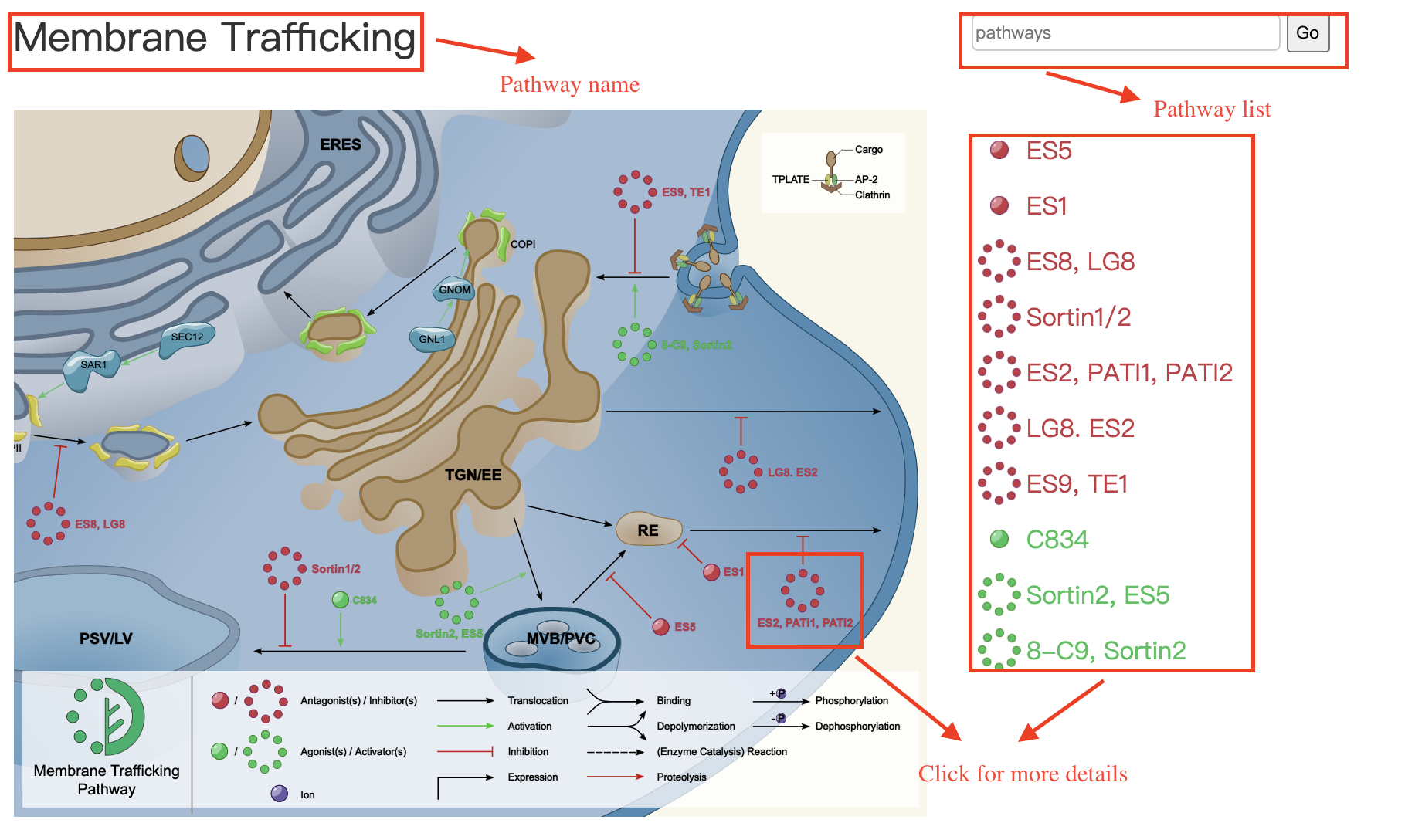
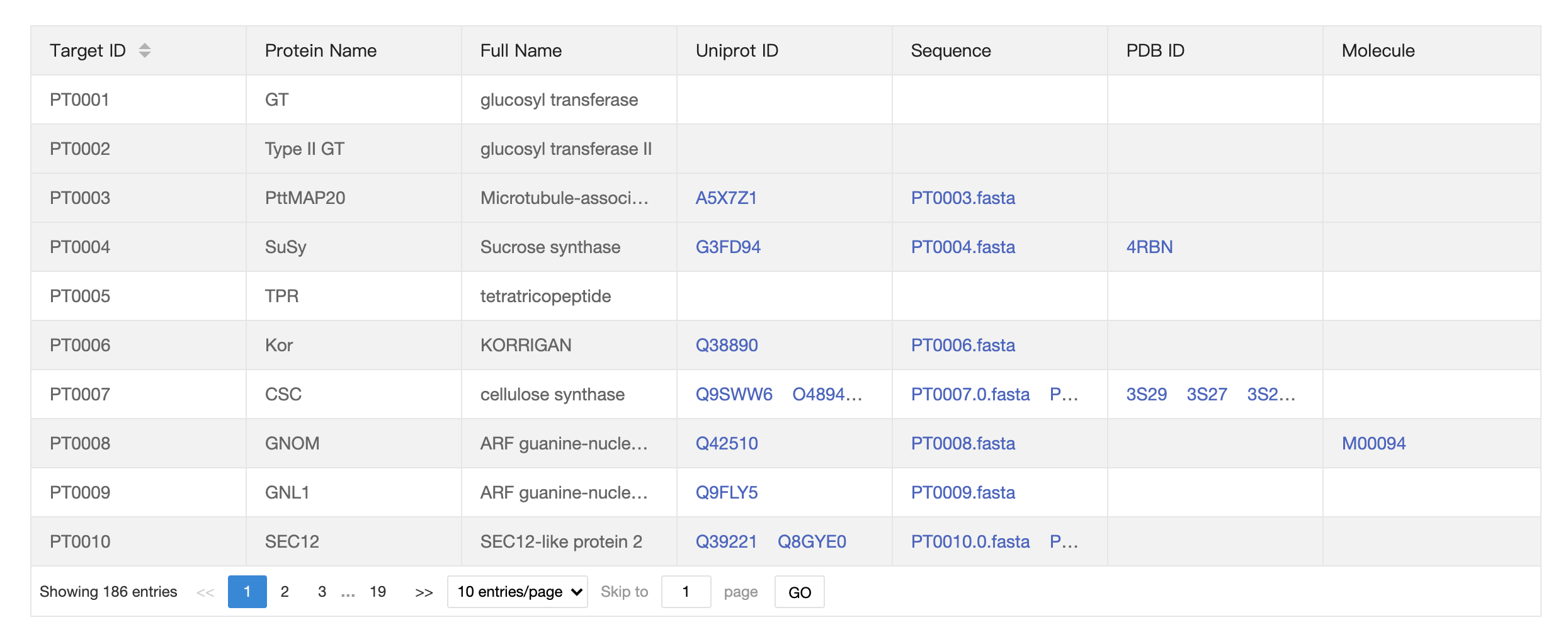
Browse-Target
Search
SMART can be retrieved through in text-based keyword search, in structure-based similarity search or in sequence-based similarity search. For text search, moleculars match the keywords will be showed. For structure search, molecules with same or similar structure with input structure will be present. For sequence search, proteins with similar sequence with input will be present.Such database searchers enable users to more deeply understand small molecules/proteins/biological processes/phyenotypes of interested.
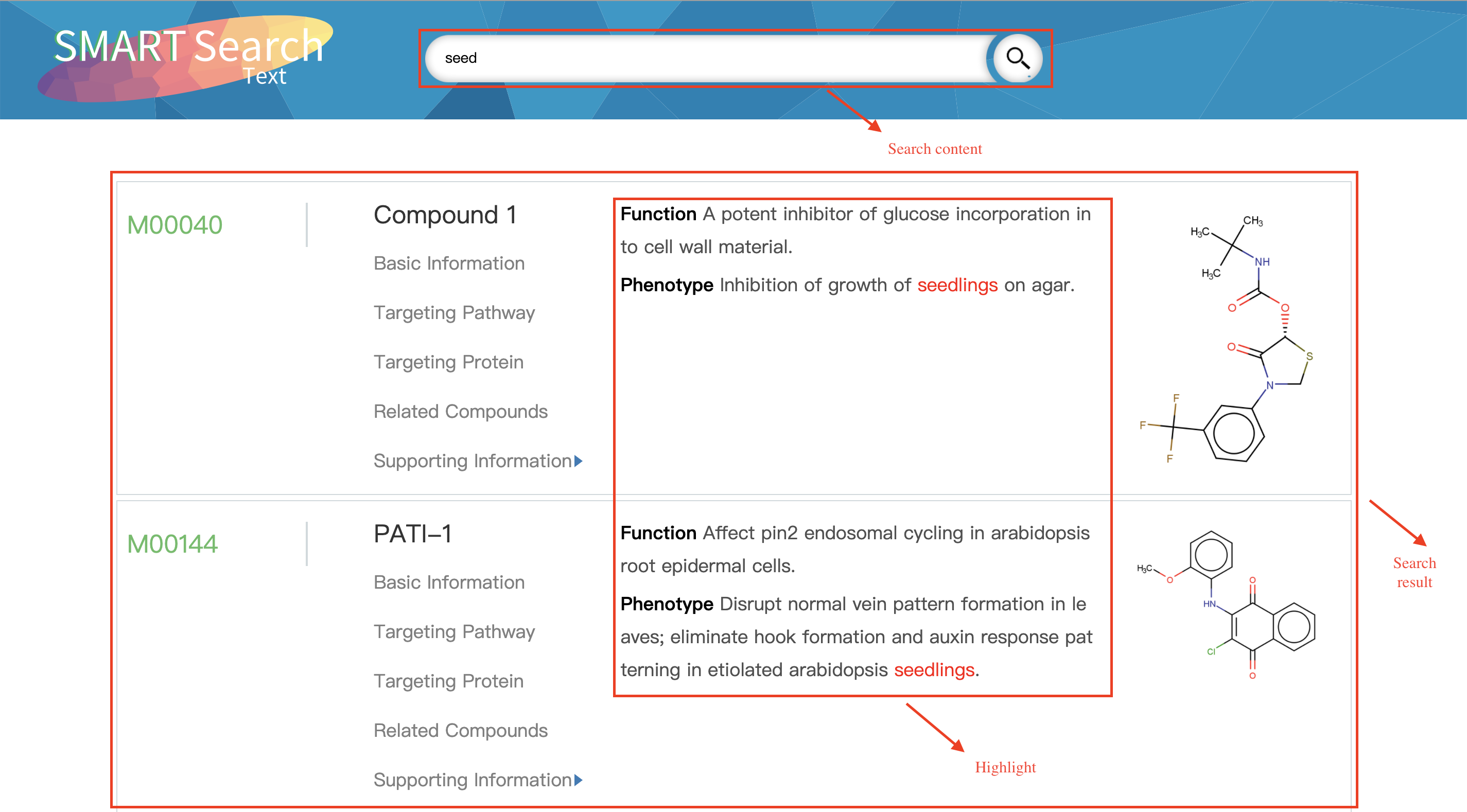
Search-Text
SMRAT supports the search for a keyword to get small molecules like the IUPAC and common name, chemical formula,phenotype information and so on. And the search can also be preformed based on target proteins (if users interested) with some keywords like protein name, uniport ID, PDB ID.
Input:Molecular name, chemical formula, phenotype keywords, etc.
Output:Molecular information
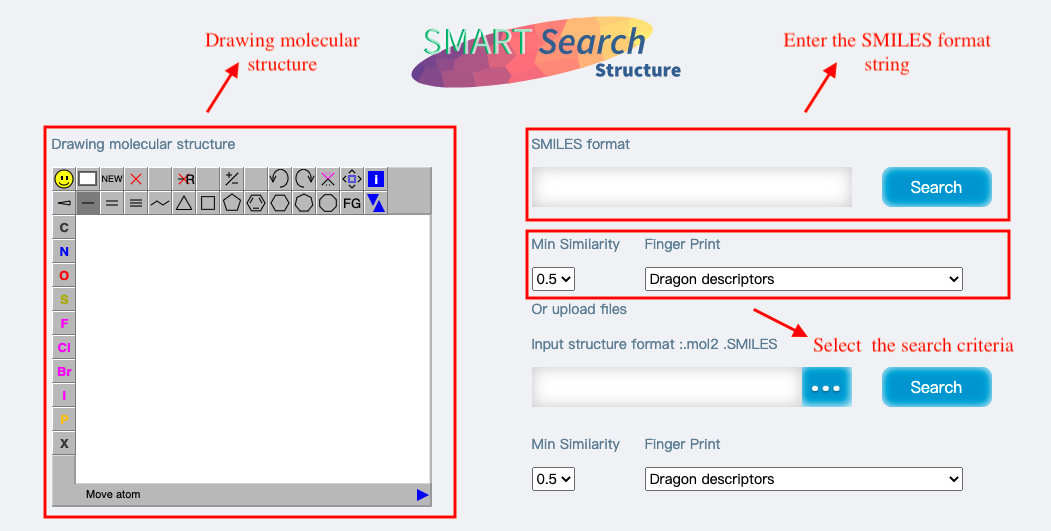
Search-Structure
SMART supports the search for 2D and 3D structures to get similar small molecules.
Input:1)Drawing the molecular structure in the JSME molecular editor. 2)Enter the SMILES format string. 3)Directly upload the molecular structure in mol2 or SMILE format,.
Output:Similar molecular information
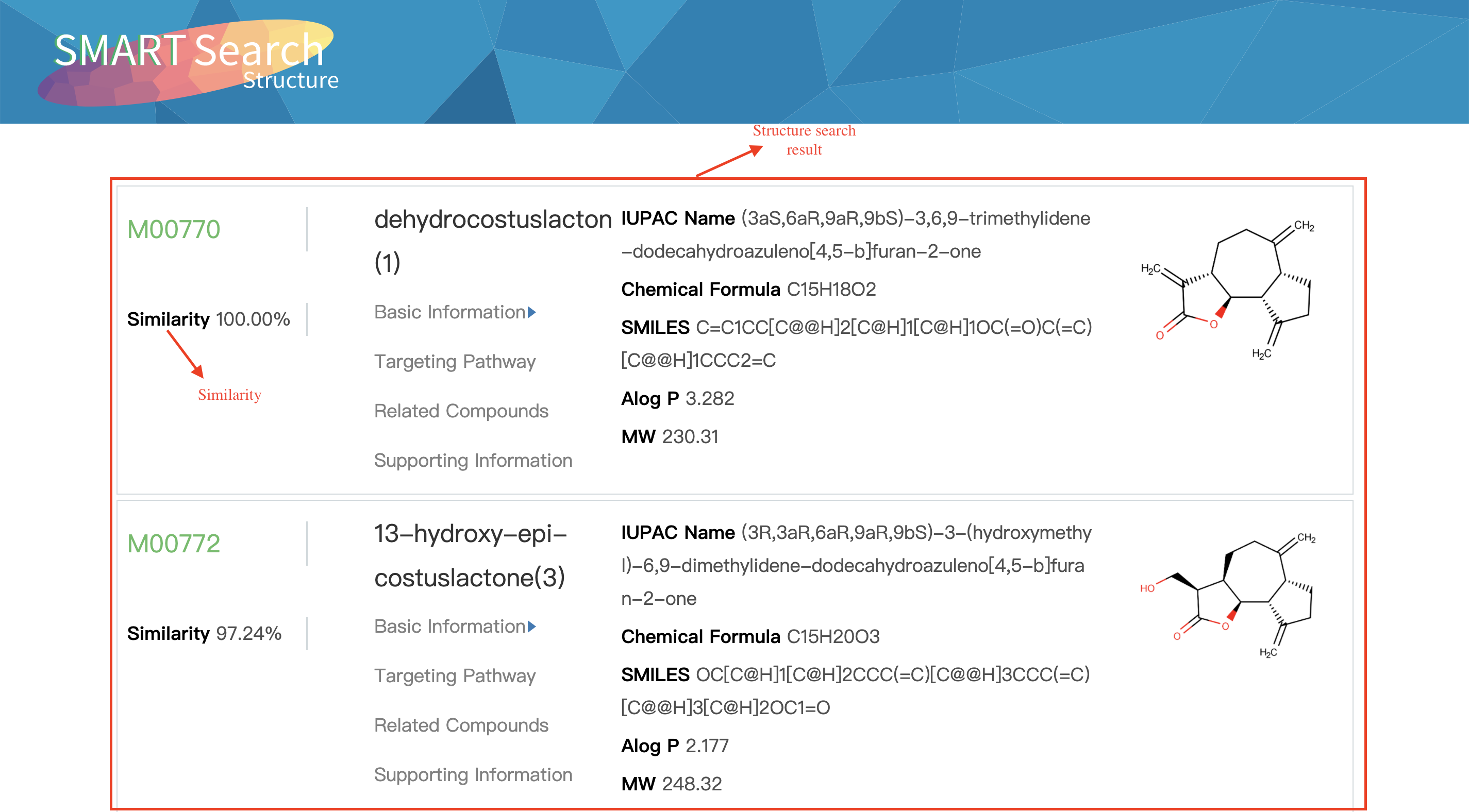
Search-Sequence
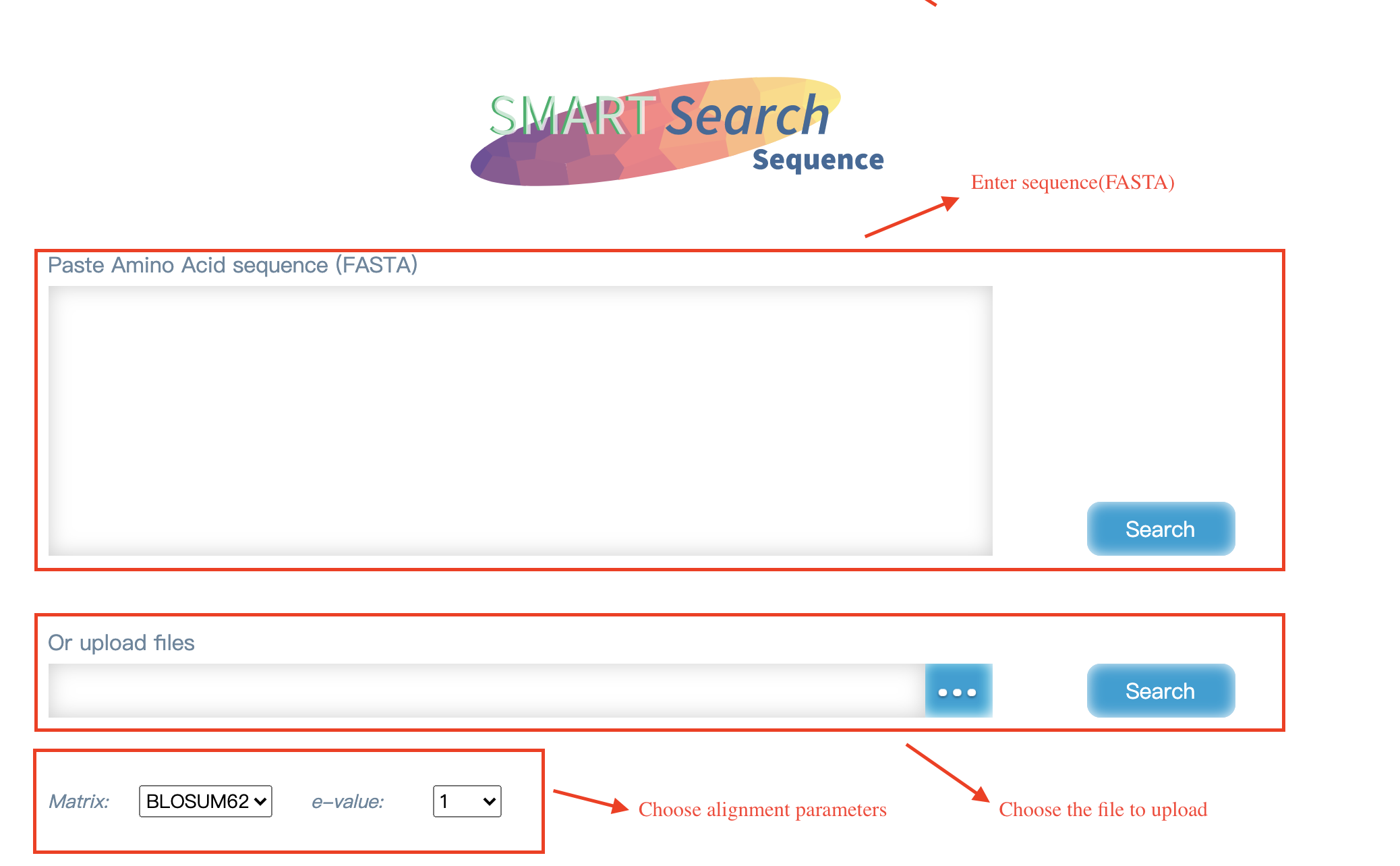
SMART supports the search for protein sequences.
Input:1)Enter the protein sequence on the page. 2)Protein sequences in FASTA format.
Output:The results comparing with protein sequences in the database by blastp.
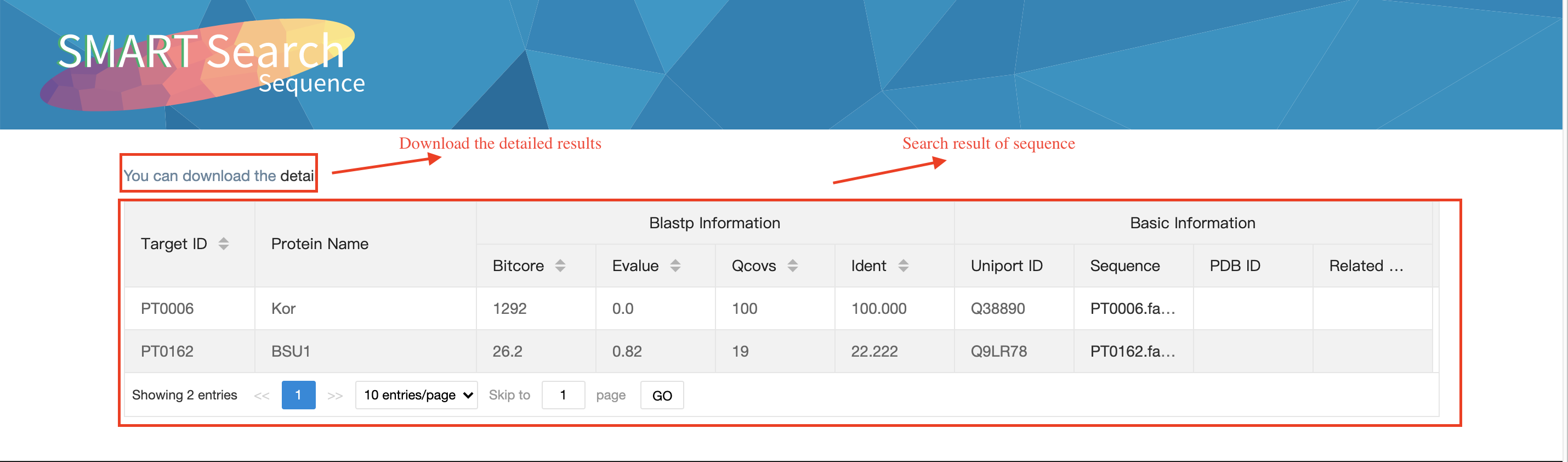
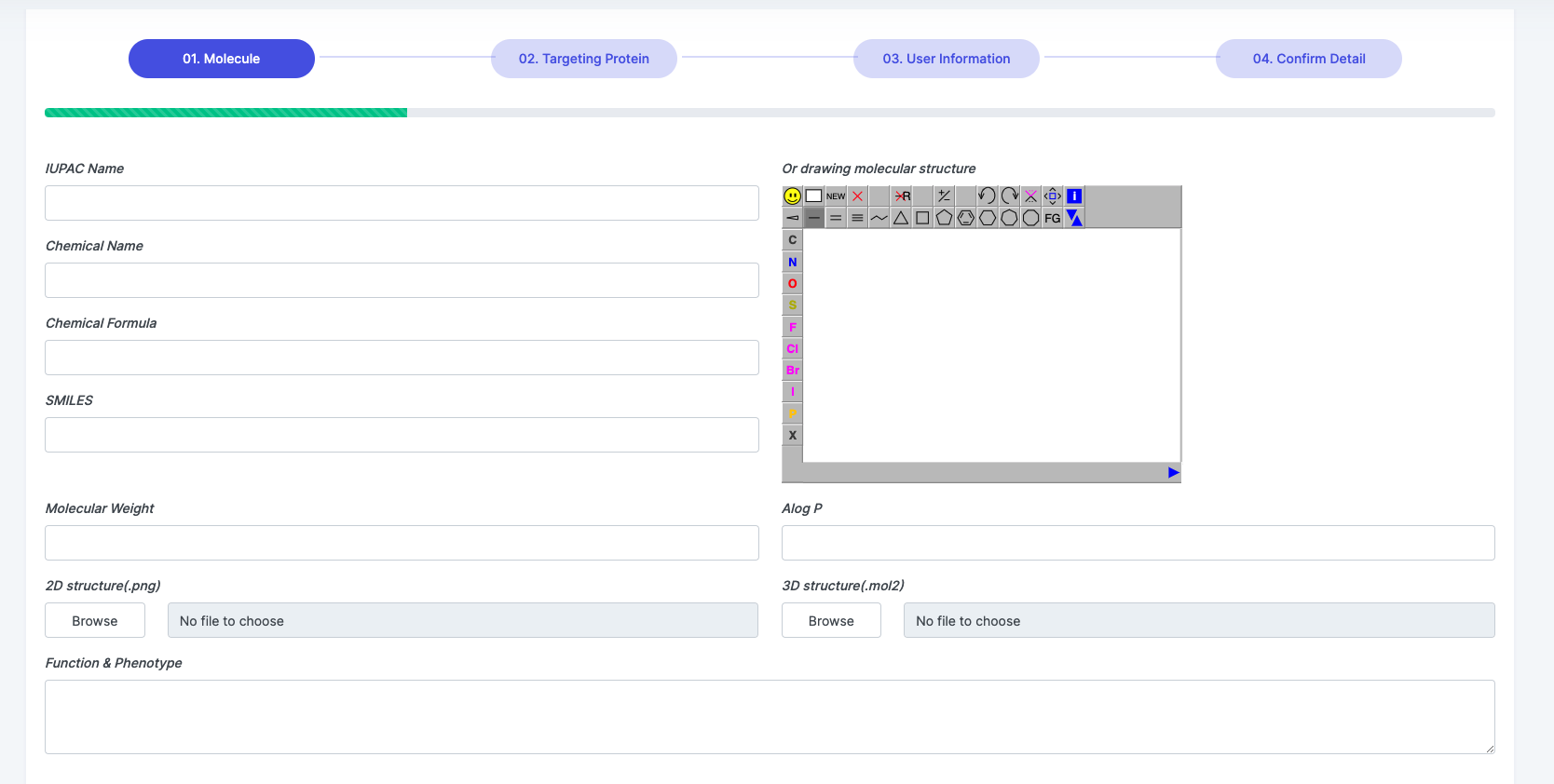
Contribute
We welcome the curation from users for any error of supplement, it can be achieved by "Correction -> Edit" at each entry webpages shown in figure below.

By clicking "Edit", the page will go to a page in which all the information can be edited as following.
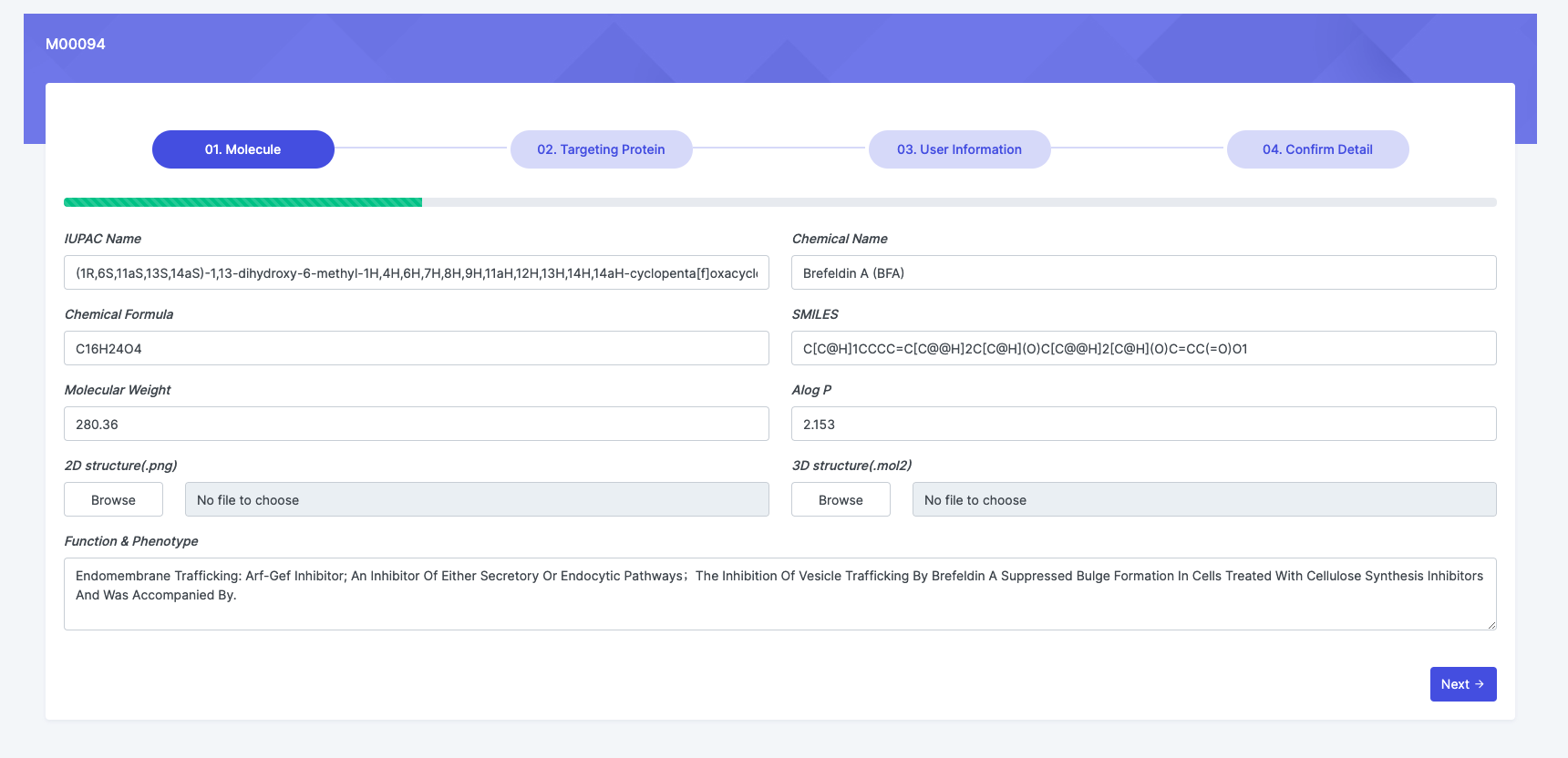
Users are also encouraged to upload their newly discovered new small molecules to the SMART database by filling the respective tables with molecular name, molecular structure, target and function informatrion.