Abstract
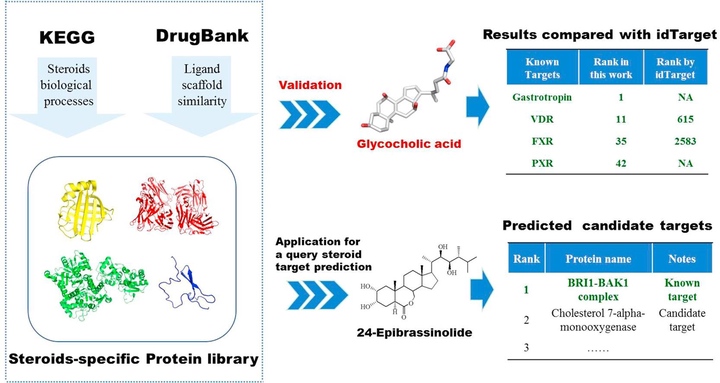
Steroids exist universally and play critical roles in various biological processes. Identifying potential targets of steroids is of great significance in studying their physiological and biochemical activities, the side effects and for drug repurposing. Herein, aiming at more precise steroids targets prediction, a steroids-specific target library integrating 3325 PDB or homology modeling structures categorized into 196 proteins was built by considering chemical similarity from DrugBank and biological processes from KEGG. The main properties of this library include: (1) It was manually prepared and checked to eliminate mistakes. (2) The library enriched the possible steroids targets and could decrease the false positives of structure-based target screening for steroids. (3) The ranking by protein name instead of PDB ID could make the screening more efficiency and precise. (4) Protein flexibility was taken into account partially by the different active conformations through the structural redundancy of each category of protein, which leads to more accurate prediction. The case studies of glycocholic acid and 24-epibrassinolide proved its powerful predictive accuracy. In summary, our strategy to build the steroids-specific protein library for steroids target prediction is a promising approach and it provides a novel idea for the target prediction of small molecules.
- 甾体特异性靶标库的构建及筛选研究
甾体类分子是生物界出现最广泛的成分之一,在各种生物过程中发挥着关键作用。确定甾体类分子的潜在靶点对研究其生理生化活性、毒副作用及生理作用具有重要意义。传统的靶标发现依赖实验,但前期的虚拟筛选能够排除大量的不可能的靶标,降低实验盲目性。现有的综合性的靶标虚拟筛选平台如idTarget和TarFishDock 包含大量晶体,给出的筛选结果为PDB晶体的列表,存在着假阳性的和蛋白结构冗余的问题。为此本课题设计了更精确的甾体类分子靶标预测方法,利用配体的化学结构相似性和KEGG的生化反应中酶和底物相互作用的原理分别从Drugbank和KEGG搜集构建了一个包含3325 晶体结构,196类蛋白的甾体特异性靶标库。该库的所有蛋白都经过人工检查以消除错误。并在前期搜集过程中考虑了甾体分子的特异性,降低了基于结构的靶标筛选的假阳性。最后的筛选结果用蛋白种类代替PDB ID进行排序,能有效提高筛选的效率和准确性。此外通过每一类蛋白的结构冗余,将不同的活性构象分数整合,能考虑到蛋白质的柔韧性,使预测更加准确。通过对甘氨胆酸和24-epibrassinolide的实例研究,发现其能有效预测出甘氨胆酸所有已知晶体结构的靶标,且与idTarget相比预测精度更高,证明了其强大的预测准确性。构建的甾体特异性蛋白库可用于对所有甾体类分子进行靶标筛选和预测,为后续的实验验证提供指导。甾体特异性靶标库的构建及后续验证比较与筛选研究流程如图3所示,相关成果已在Steroids(Dang et al.,2018)发表。